Bacteria live pretty much everywhere, dividing happily in places from stomach acid to deep-sea vents. Bacteria and Archaea can live in so many different places because their genomes are incredibly flexible–they can alter, lose and duplicate genes almost at will. Although scientists knew that prokaryotes could acquire genes from their neighbors (it’s a known contributor to antibiotic resistance), this method of gaining new DNA was thought to be very rare and only occur under strong pressures in the environment, like deadly antibiotics.
The idea that bacteria could share their genes, known as horizontal gene transfer, was first discovered by Japanese microbiologists in 19591 in a study of antibiotic resistance in different species of bacteria. They found that bacteria could acquire entire genes from other bacteria–even other species of bacteria. Since then, evolutionary biologists have been trying to figure out how important this feature is to both bacteria and the evolution of life on earth.
One way to study this is to look at similarities and differences in bacterial genes in different species. Biologist Linda Fothergill-Gilmore wrote in a review paper2 that the first bacterial genome was likely small and simple. As time passed, genes got duplicated and mutated, ultimately giving rise to the complicated synthesis and breakdown of sugars, proteins, nucleic acids, and fats that we see today. Eukaryotes are only known to use this method of gene duplication and mutation as fodder for evolution. But the 1959 Japanese study, followed by thousands of paper on horizontal gene transfer, hinted that bacteria had another way of acquiring new genes. If a bacterial gene evolved in the “standard” eukaryotic way–by duplicating and mutating over time–the new gene’s function would be obviously related to other genes in the bacterium. If, on the other hand, a bacterium got a new gene by swallowing an entire gene from its neighbor (lending new meaning to the phrase “Big Gulp”), and then pass that gene along to its daughter cells (known as vertical gene transfer), that swallowed gene wouldn’t have any close links to its host genome.
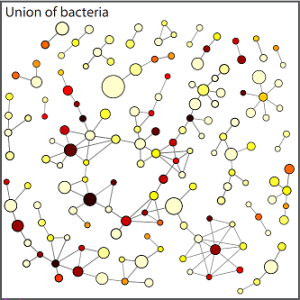
In a paper published yesterday in BMC Evolutionary Biology, a team led by Sebastian Bernhardsson of the Niels Bohr Institute created a network to show how different genes were shared amongst 134 different species of bacteria3. Bernhardsson’s team called each gene a “node” and noted how many different bacterial species shared this gene. The bigger the node (pictured as a circle), the more bacterial species that shared this gene. Their results looked like this (Figure 1):
Not surprisingly, they found a large core of basic enzymes that performed common duties in many different bacteria. Most of these genes appeared to arise in the “eukaryotic” way, by a gene getting duplicated and then mutating and doing something new. These genes were more likely to have close evolutionary links to nearby genes, which indicates that the two genes originated from the same source. At the periphery of the network, where genes were shared among relatively few bacterial species, Bernhardsson and colleagues found that bacteria were more likely to have acquired these genes through horizontal gene transfer. Obtaining a more peripheral gene may not sound like a big deal, but it likely gave the bacteria radically new functionality. And new functionality means the bacteria can live in a new environment and evolve further.
This result, the authors write, “lends additional support to the importance of horizontal gene transfer during bacterial metabolic evolution where new reactions are attached at the periphery of the network.”
Whereas horizontal gene transfer may not have had a significant effect early in the evolution of metabolism, it has allowed bacteria to expand their abilities into new areas later in the history of life.
This pup summarizes the results thusly:

References:
1. Ochiai K, Yamanaka T, Kimura K, Sawada, O (1959). “Inheritance of drug resistance (and its transfer) between Shigella strains and Between Shigella and E. coli strains” (in Japanese). Hihon Iji Shimpor 1861: 34
2. Fothergill-Gilmore LA (1986). “The evolution of the glycolytic pathway.” Trends in Biochemical Sciences 11:47-51. doi:10.1016/0968-0004(86)90233-1
3. Bernhardsson S, Gerlee P, Linzana L (2011). “Structural correlations in bacterial metabolic networks.” BMC Evolutionary Biology. doi:10.1186/1471-2148-11-20